Research
Dissecting cell-cell interactions in the tumour microenvironment (TME)
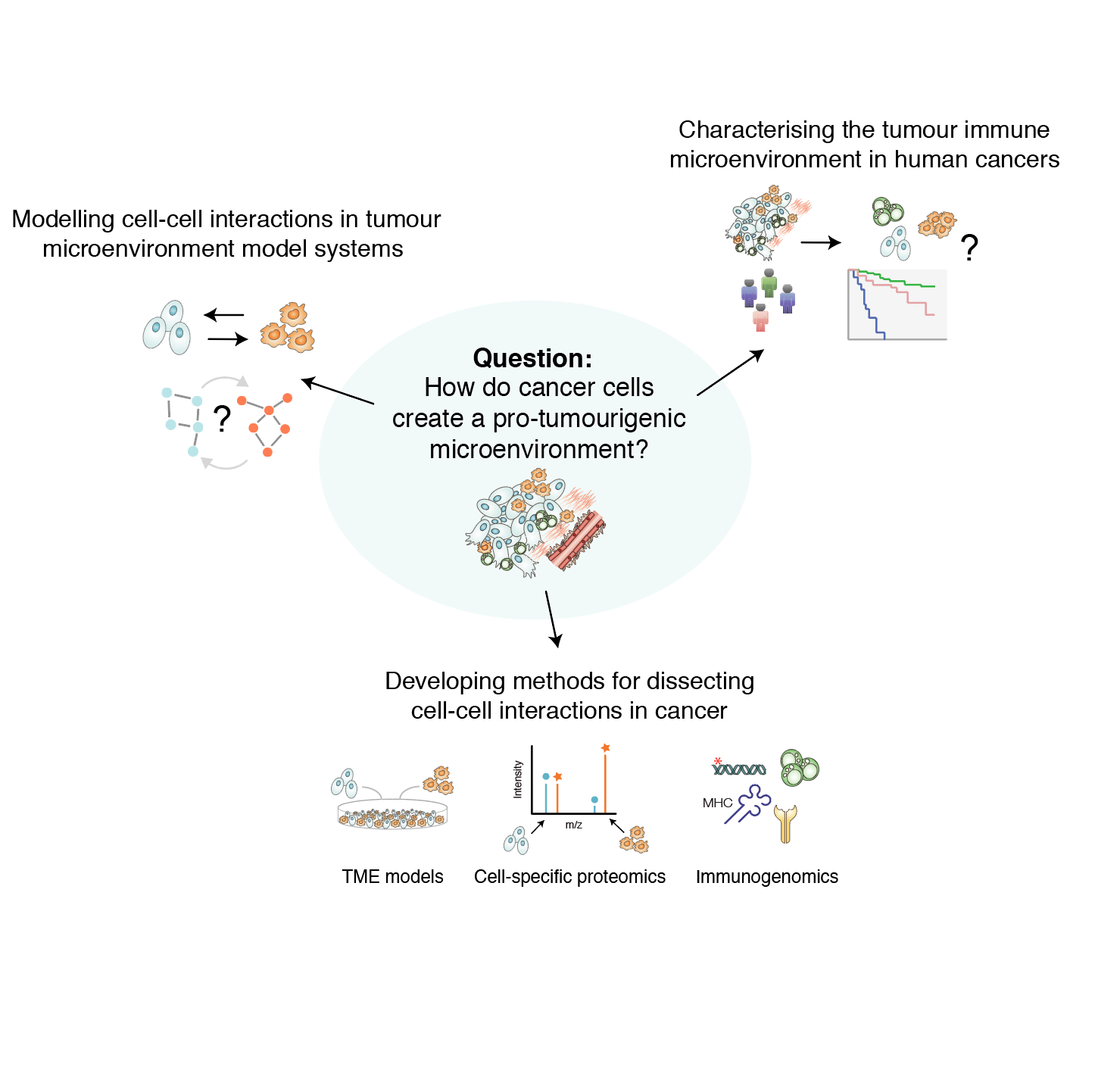
We are a cancer systems biology group that uses experimental and computational approaches to investigate the tumour microenvironment. The fundamental question that drives our research is: How do cancers create a pro-tumourigenic microenvironment? To decipher intercellular communication in cancer, we develop and apply technologies that can decompose the transcriptome or label the proteome of specific cell populations in multicellular settings. Our long-term research goal is to better understand how immune cells and stromal cells are involved in oncogenesis and how this can be exploited in cancer therapy.
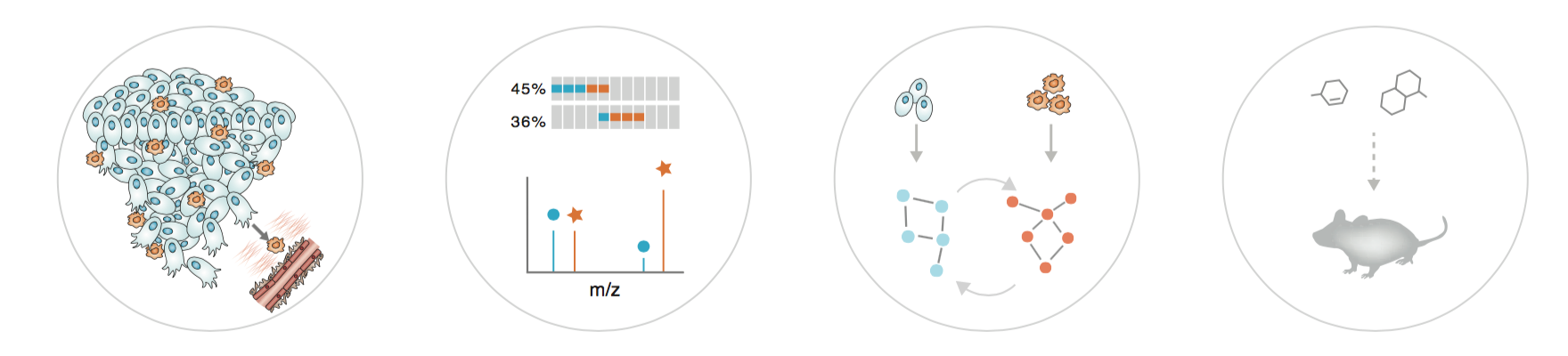
Characterising the TME in human cancers: Using immunogenomics analysis of a patient case with metastatic ovarian cancer, we have recently shown how tumours within the same individual can have vastly different immune microenvironments, and moreover, that immune cell infiltration and exclusion associate with tumour regression and progression, respectively (Jiménez-Sánchez et al 2017, Cell). In addition, we have identified that oncogenic Wnt/β-catenin and Myc signalling associate with immune exclusion and that chemotherapy induces immunogenic responses in HGSOC (Jiménez-Sánchez et al 2020, Nat Genet). To further investigate the tumour microenvironment across human cancer types, we take an integrative approach combining data from publicly available clinical data with data from cancer genomics (TCGA) and proteomics studies (CPTAC). We perform functional analyses of associations between clinical features, cell infiltrations, and somatic mutations in signaling pathways and protein domains (see e.g. Ciriello et al 2013, Nat Genet; Miller et al 2015, Cell Syst; Gauthier et al 2016, NAR; Jimenez-Sanchez, Cast, Miller 2019, Cancer Res). Through our on-going collaborations with clinician-scientists, we will also characterise the tumour-immune microenvironment of primary and metastatic tumour samples.
Cell-cell communication in the TME: In many human cancer types, stromal cells in the tumour microenvironment are causally involved in cancer development and resistance to therapy. We have recently developed a new proteomic method (CTAP) to globally interrogate the cell-of-origin of proteins in cells in coculture, greatly facilitating studies of cell-to-cell communication in models of the tumour microenvironment (Gauthier et al 2013, Nat Methods). We will use CTAP and quantitative mass spectrometry to systematically characterise tumour-stroma interactions in order to identify effective drug treatments that overcome both tumour intrinsic and -extrinsic mechanisms of drug resistance. Combining cell-selective proteomic methods with pathway analysis and perturbation-based network modeling (Molinelli et al 2013, PLOS Comp Biol; Miller et al 2013, Sci Sig), our research will pioneer modelling of cell-cell communication in the tumor microenvironment of cancers such as pancreatic ductal adenocarcinoma (PDAC).
Please visit our group webpage at the CRUK, Cambridge Institute.