Webservices
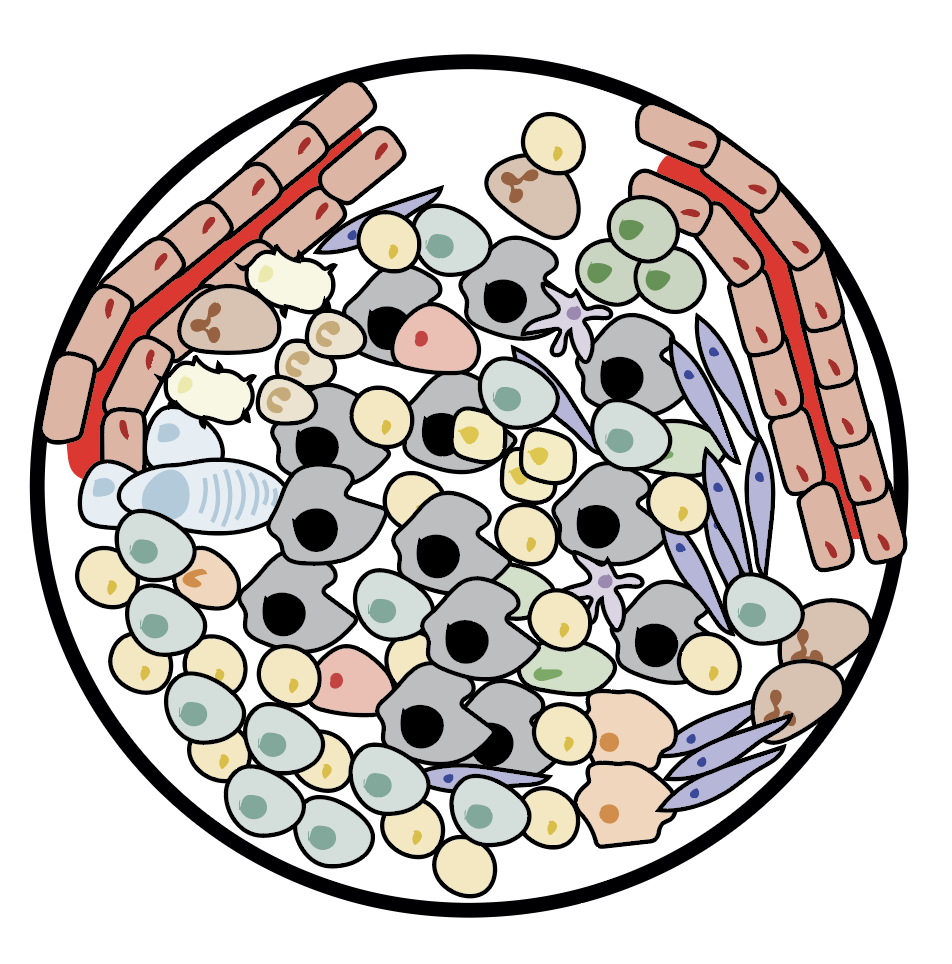
ConsensusTME
ConsensusTME is a resource for estimation of immune cell composition of tumour samples using bulk mRNA data (Jimenez-Sanchez, Cast and Miller, 2019, Cancer Research)
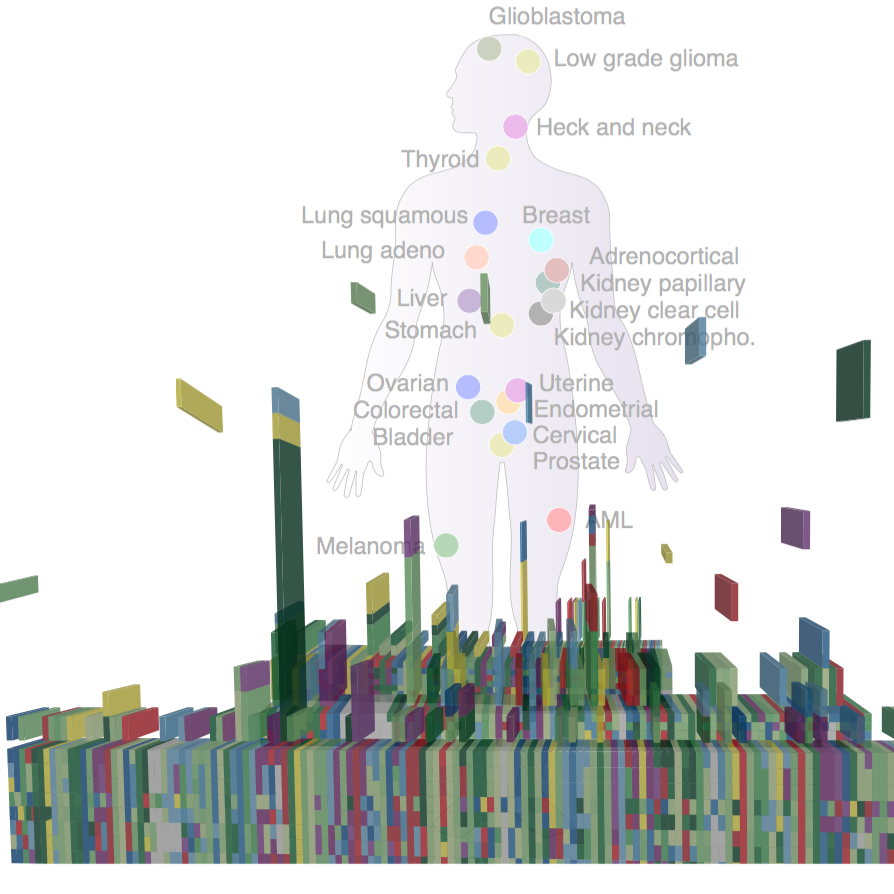
MutationAligner [with MSKCC]
MutationAligner is a resource that enables discovery and exploration of somatic mutation hotspots identified in protein domains in more than 5000 cancer patient samples across 22 different tumor types (Miller et al, 2015, Cell Systems, Gauthier et al, 2015, Nucleic Acids Res)
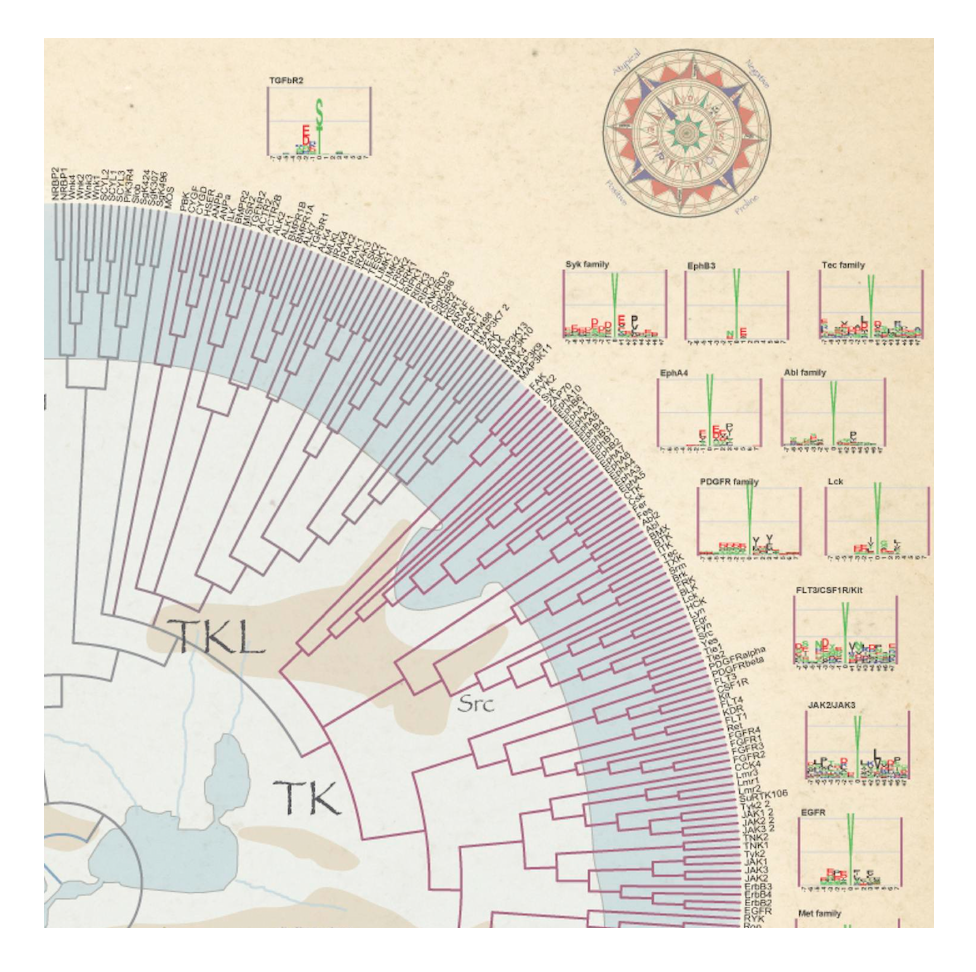
Netphorest [with UCPH]
Netphorest is a resource of 125 sequence-based classifiers of linear motifs in phosphorylation-dependent signaling with coverage of 179 of 518 kinases, & 93 of 118 SH2 domains, 8 of 18 PTB domains, BRCT and WW domains (Miller et al, 2008, Science Signaling; Horn et al, 2014, Nature Methods)
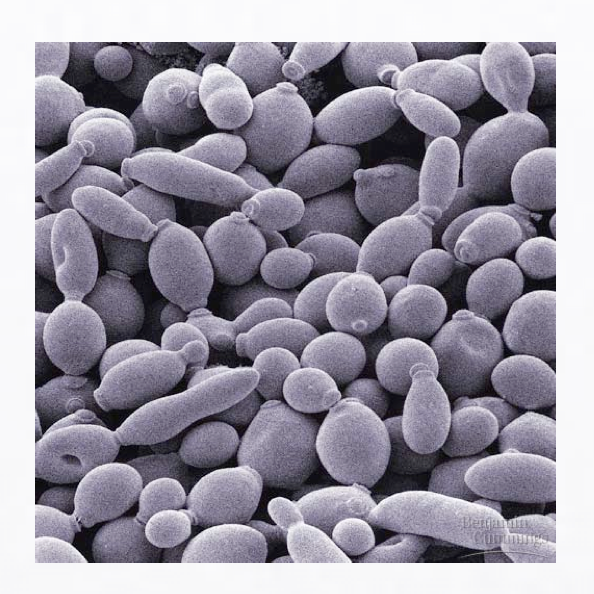
NetPhosYeast [with DTU]
A webservice for prediction of generic protein phosphorylation sites in yeast (Ingrell et al 2007, Bioinformatics)
NetPhosBac [with DTU]
A webservice for prediction of generic protein phosphorylation sites in bacteria (Miller et al, 2008, Proteomics)